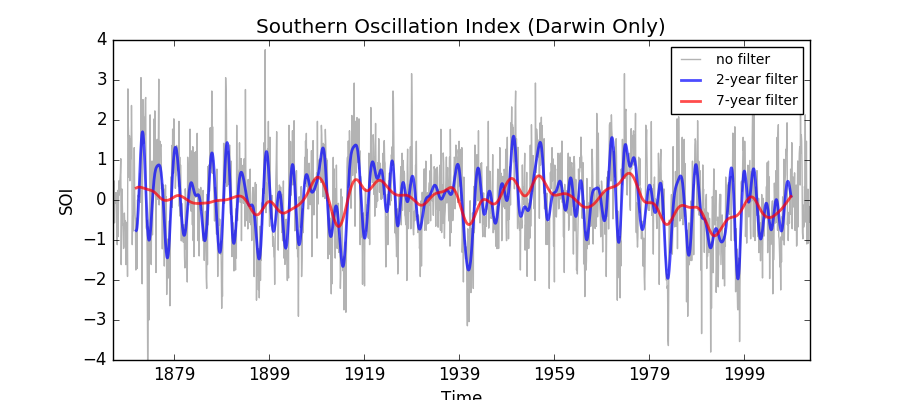
This example demonstrates low pass filtering a time-series by applying a weighted running mean over the time dimension.
The time-series used is the Darwin-only Southern Oscillation index (SOI), which is filtered using two different Lanczos filters, one to filter out time-scales of less than two years and one to filter out time-scales of less than 7 years.
Duchon C. E. (1979) Lanczos Filtering in One and Two Dimensions. Journal of Applied Meteorology, Vol 18, pp 1016-1022.
Trenberth K. E. (1984) Signal Versus Noise in the Southern Oscillation. Monthly Weather Review, Vol 112, pp 326-332
"""
Applying a filter to a time-series
==================================
This example demonstrates low pass filtering a time-series by applying a
weighted running mean over the time dimension.
The time-series used is the Darwin-only Southern Oscillation index (SOI),
which is filtered using two different Lanczos filters, one to filter out
time-scales of less than two years and one to filter out time-scales of
less than 7 years.
References
----------
Duchon C. E. (1979) Lanczos Filtering in One and Two Dimensions.
Journal of Applied Meteorology, Vol 18, pp 1016-1022.
Trenberth K. E. (1984) Signal Versus Noise in the Southern Oscillation.
Monthly Weather Review, Vol 112, pp 326-332
"""
import numpy as np
import matplotlib.pyplot as plt
import iris
import iris.plot as iplt
def low_pass_weights(window, cutoff):
"""Calculate weights for a low pass Lanczos filter.
Args:
window: int
The length of the filter window.
cutoff: float
The cutoff frequency in inverse time steps.
"""
order = ((window - 1) // 2) + 1
nwts = 2 * order + 1
w = np.zeros([nwts])
n = nwts // 2
w[n] = 2 * cutoff
k = np.arange(1., n)
sigma = np.sin(np.pi * k / n) * n / (np.pi * k)
firstfactor = np.sin(2. * np.pi * cutoff * k) / (np.pi * k)
w[n-1:0:-1] = firstfactor * sigma
w[n+1:-1] = firstfactor * sigma
return w[1:-1]
def main():
# Enable a future option, to ensure that the netcdf load works the same way
# as in future Iris versions.
iris.FUTURE.netcdf_promote = True
# Load the monthly-valued Southern Oscillation Index (SOI) time-series.
fname = iris.sample_data_path('SOI_Darwin.nc')
soi = iris.load_cube(fname)
# Window length for filters.
window = 121
# Construct 2-year (24-month) and 7-year (84-month) low pass filters
# for the SOI data which is monthly.
wgts24 = low_pass_weights(window, 1. / 24.)
wgts84 = low_pass_weights(window, 1. / 84.)
# Apply each filter using the rolling_window method used with the weights
# keyword argument. A weighted sum is required because the magnitude of
# the weights are just as important as their relative sizes.
soi24 = soi.rolling_window('time',
iris.analysis.SUM,
len(wgts24),
weights=wgts24)
soi84 = soi.rolling_window('time',
iris.analysis.SUM,
len(wgts84),
weights=wgts84)
# Plot the SOI time series and both filtered versions.
plt.figure(figsize=(9, 4))
iplt.plot(soi, color='0.7', linewidth=1., linestyle='-',
alpha=1., label='no filter')
iplt.plot(soi24, color='b', linewidth=2., linestyle='-',
alpha=.7, label='2-year filter')
iplt.plot(soi84, color='r', linewidth=2., linestyle='-',
alpha=.7, label='7-year filter')
plt.ylim([-4, 4])
plt.title('Southern Oscillation Index (Darwin Only)')
plt.xlabel('Time')
plt.ylabel('SOI')
plt.legend(fontsize=10)
iplt.show()
if __name__ == '__main__':
main()
(Source code, png)